What are we doing?

Shewanella
Our group is mainly using species of the genus Shewanella as model organisms to study various aspects of the bacteria in general. Shewanella sp. are versatile facultatively anaerobic gammaproteobacteria that can thrive in a wide range of environmental conditions and are capable of using an impressive array of alternative terminal electron acceptors, including numerous metal oxides. Shewanella species have potential to be used in different bioremediation processes and applications such as microbial fuel cells, but have also been identified as commensal pathogens. The numerous sequenced species that were isolated from various environments make Shewanella a well-suited model for long-term environmental adaptation. To study our fields of interest, we are using a wide range of experimental approaches. Almost all genetic tools are available for members of this genus.

Flagella
Flagella are long helical proteinaceous filaments extending from the cell’s surface that are rotated at the filaments base by a membrane-embedded motor. The flagellar motor is a highly intricate nanomachine which is powered by transmembrane ion gradients. Most bacterial motors interact with one or more corresponding chemotaxis systems to convert the perception of environmental gradients of attractants or repellents into directed movement. Production and placement of the huge flagellar and chemotaxis machinery requires tight spatiotemporal control and thus serve as great models to study the cellular organization of bacterial cells.
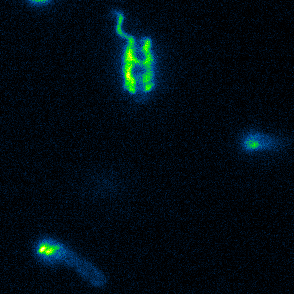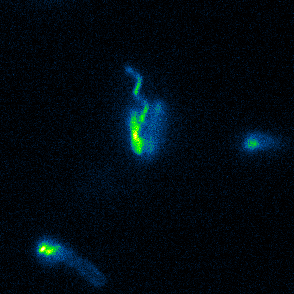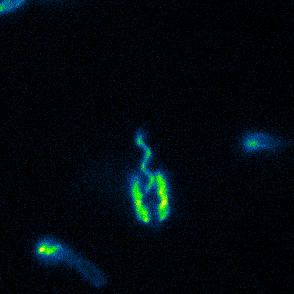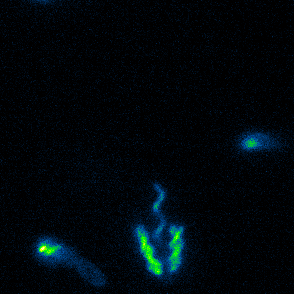
Aspects of flagellar function and cellular navigation
Our main Shewanella model species, S. putrefaciens CN-32, possesses
two complete flagellar systems which are synchronously assembled in a subpopulation of the cells. The primary system forms the sodium ion-dependent single polar flagellum typical for
Shewanella sp., the second system produces one or more lateral flagellar filaments. S. putrefaciens can also perform different modes of flagella-mediated motility, regular free
swimming with the flagellar filament pointing away from the cell and another mode, where the polar flagellar filament is wrapped around the cell body. This formation can be used for so-called
screw thread motility to glide across surfaces and, by this, escape from contricted environments.
In various projects we are investigating further how bacterial cells move through structured environments, how the flagellar filaments function and how flagellar activity is regulated.

Cell polarity
Spatiotemporal organization is crucial for a number of fundamental cell processes such as cell division or motility. In S. putrefaciens, the primary flagellar system and the associated flagellar systems are localized to the cell pole, while the secondary flagellar system is not. Recruitment of one, but not the other complex flagellar machinery to a specific location within the cell, as in S. putrefaciens CN-32, provides an excellent system to study the underlying mechanisms of regulation and flagellar placement which is conducted in close collaboration with the lab of Gert Bange in Marburg.
In many bacterial species, number and position of flagella depend on two proteins, the SRP-type GTPase FlhF and the MinD-like ATPase FlhG (also named YlxH, MinD2, FleN or MotR). In S. putrefaciens CN-32, loss of one or both proteins results in an aberrant flagellation pattern of the primary, but not the secondary flagellar system. We could show that for proper function, FlhG needs to interact with building blocks of the flagellar basal body, namely FliM and N. The corresponding binding motif is absent in the secondary flagellar system which is therefore not addressed by FlhG. Thus, FlhG links regulation and assembly of a multiprotein complex. We are currently further investigating the function and mechanism by which the two highly similar proteins mediate a wide range of different flagellation patterns in various bacterial species.

Phage biology
The viruses of bacteria, the phages, are extremely abundant and are thought to outnumber their hosts by a factor of ten. Over billions of years, phages and bacteria are engaged in a race-of-arms and affect each other and their populations in various ways we are only beginning to understand. The advent of advanced techniques in e.g. molecular biology, proteinbiochemistry, microscopy and next-generation sequencing and the identification of a huge number of novel phages has lead to a new wave of research on phages. The studies are ranging from the molecular mechanisms of phage-host interactions to various potential applications of phages and their proteins, e.g. as an addition to antibiotic treatments. We are interested in novel aspects of phage-host interactions.
